Sinkhorn Divergence Hessians
Contents
Sinkhorn Divergence Hessians#
Samples two point clouds, computes their Sinkhorn divergence#
We show in colab how OTT and JAX can be used to compute automatically the Hessian of the sinkhorn_divergence() w.r.t. the input variables, such as weights a or locations x.
import sys
if "google.colab" in sys.modules:
!pip install -q git+https://github.com/ott-jax/ott@main
import jax
import jax.numpy as jnp
import matplotlib.pyplot as plt
from ott.geometry import pointcloud
from ott.solvers.linear import implicit_differentiation as implicit_lib
from ott.tools import sinkhorn_divergence
Sample two random point clouds of dimension dim
def sample(n, m, dim):
rngs = jax.random.split(jax.random.PRNGKey(0), 6)
x = jax.random.uniform(rngs[0], (n, dim))
y = jax.random.uniform(rngs[1], (m, dim))
a = jax.random.uniform(rngs[2], (n,)) + 0.1
b = jax.random.uniform(rngs[3], (m,)) + 0.1
a = a / jnp.sum(a)
b = b / jnp.sum(b)
return a, x, b, y
a, x, b, y = sample(15, 17, 3)
As usual in JAX, we define a custom loss that outputs the quantity of interest, and is defined using relevant inputs as arguments, i.e. parameters against which we may want to differentiate. We add to a and x the implicit auxiliary flag which will be used to switch between unrolling and implicit differentiation of the Sinkhorn algorithm (see this excellent tutorial for a deep dive on their differences).
The loss outputs the Sinkhorn divergence between two point clouds.
def loss(a, x, implicit):
return sinkhorn_divergence.sinkhorn_divergence(
pointcloud.PointCloud,
x,
y, # this part defines geometry
a=a,
b=b, # this sets weights
sinkhorn_kwargs={
"implicit_diff": implicit_lib.ImplicitDiff(
precondition_fun=lambda x: x
),
"use_danskin": False,
}, # to be used by the Sinkhorn algorithm
).divergence
Let’s parse the three lines in the call to sinkhorn_divergence() above:
The first one defines the point cloud geometry between
xandythat will define the cost matrix. Here we could have added details onepsilonregularization (or scheduler), as well as alternative definitions of the cost function (here assumed by default to be squared Euclidean distance). We stick to the default setting.The second one sets the respective weight vectors
aandb. Those are simply two histograms of sizenandm, both sum to 1, in the so-called balanced setting.The third one passes on arguments to the three
Sinkhornsolvers that will be called, to comparexwithy,xwithxandywithywith their respective weightsaandb. Rather than focusing on the several numerical options available to parameterizeSinkhorn’s behavior, we instruct JAX on how it should differentiate the outputs of the sinkhorn algorithm. Theuse_danskinflag specifies whether the outputted potentials should be freezed when differentiating. Since we aim for 2nd order differentiation here, we must set this toFalse(if we wanted to compute gradients,Truewould have resulted in faster yet almost equivalent computations).
Computing Hessians#
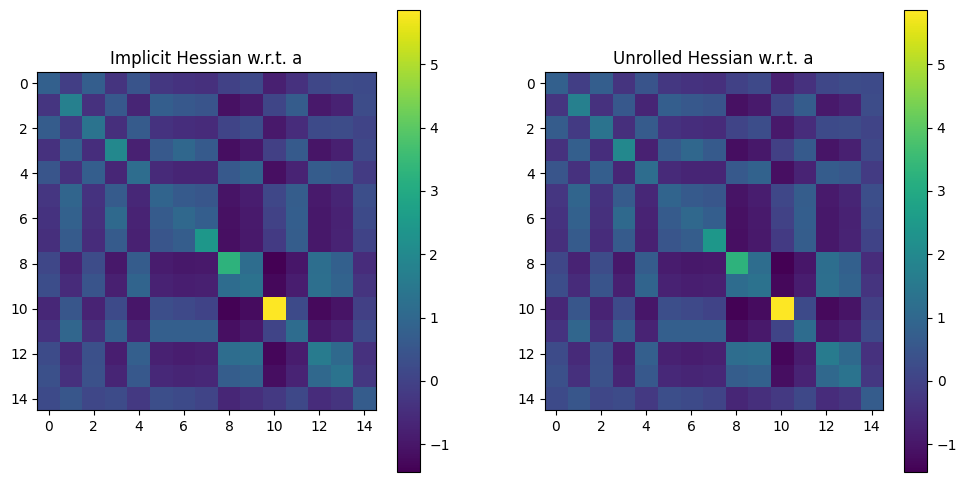
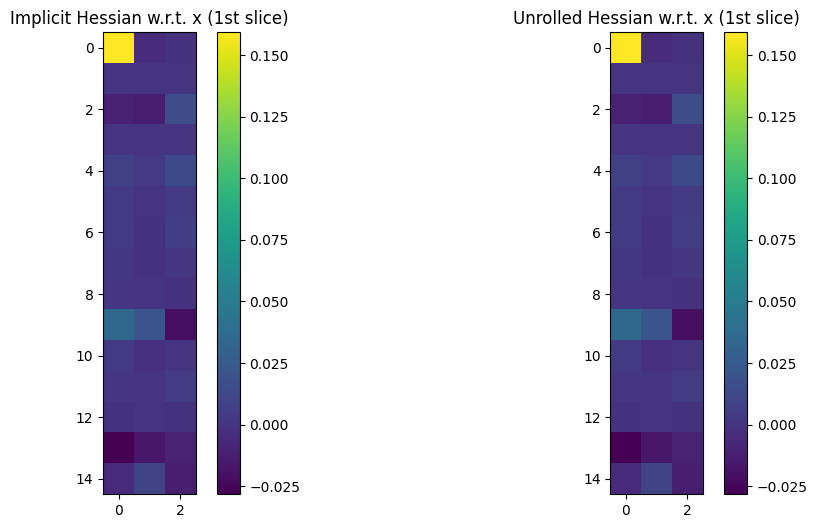
Let’s now plot Hessians of this output w.r.t. either a or x.
The Hessian w.r.t.
awill be a \(n \times n\) matrix, with the convention thatahas size \(n\).Because
xis itself a matrix of 3D coordinates, the Hessian w.r.t.xwill be a 4D tensor of size \(n \times 3 \times n \times 3\).
To plot both Hessians, we loop on arg 0 or 1 of loss, and plot all (or part for x) of those Hessians, to check they match:
for arg in [0, 1]:
# Compute Hessians using either unrolling or implicit differentiation.
hess_loss_imp = jax.jit(
jax.hessian(lambda a, x: loss(a, x, True), argnums=arg)
)
print("--- Time: Implicit Hessian w.r.t. " + ("a" if arg == 0 else "x"))
%timeit _ = hess_loss_imp(a, x).block_until_ready()
hess_imp = hess_loss_imp(a, x)
hess_loss_back = jax.jit(
jax.hessian(lambda a, x: loss(a, x, False), argnums=arg)
)
print("--- Time: Unrolled Hessian w.r.t. " + ("a" if arg == 0 else "x"))
%timeit _ = hess_loss_back(a, x).block_until_ready()
hess_back = hess_loss_back(a, x)
# Since we are solving balanced OT problems, Hessians w.r.t. weights are
# only defined up to the orthogonal space of 1s.
# For that reason we remove that contribution and check the
# resulting matrices are equal.
if arg == 0:
hess_imp -= jnp.mean(hess_imp, axis=1)[:, None]
hess_back -= jnp.mean(hess_back, axis=1)[:, None]
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
im = ax1.imshow(hess_imp if arg == 0 else hess_imp[0, 0, :, :])
ax1.set_title(
"Implicit Hessian w.r.t. " + ("a" if arg == 0 else "x (1st slice)")
)
fig.colorbar(im, ax=ax1)
im = ax2.imshow(hess_back if arg == 0 else hess_back[0, 0, :, :])
ax2.set_title(
"Unrolled Hessian w.r.t. " + ("a" if arg == 0 else "x (1st slice)")
)
fig.colorbar(im, ax=ax2)
--- Time: Implicit Hessian w.r.t. a
11.2 ms ± 24.1 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
--- Time: Unrolled Hessian w.r.t. a
11.2 ms ± 13.5 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
--- Time: Implicit Hessian w.r.t. x
35.2 ms ± 132 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
--- Time: Unrolled Hessian w.r.t. x
35.2 ms ± 289 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)